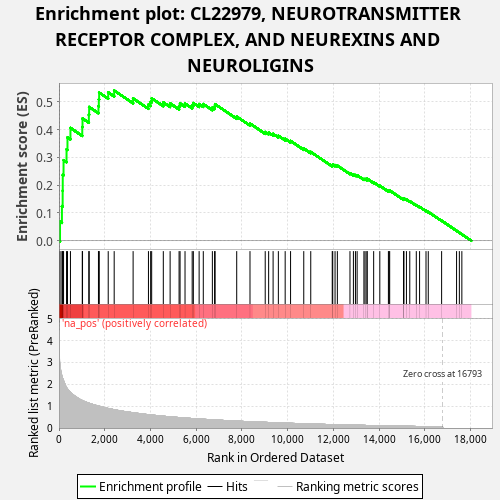
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Y98.1 Radiology of one body area (or < 20 minutes)__41210-both_sexes-Y981 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL22979, NEUROTRANSMITTER RECEPTOR COMPLEX, AND NEUREXINS AND NEUROLIGINS |
| Enrichment Score (ES) | 0.5406201 |
| Normalized Enrichment Score (NES) | 1.2807015 |
| Nominal p-value | 0.013 |
| FDR q-value | 0.3763452 |
| FWER p-Value | 0.911 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | NRXN3 | 45 | 2.815 | 0.0689 | Yes |
| 2 | MDGA1 | 128 | 2.368 | 0.1245 | Yes |
| 3 | FBXO40 | 160 | 2.255 | 0.1800 | Yes |
| 4 | SHANK1 | 164 | 2.249 | 0.2369 | Yes |
| 5 | PTCHD1 | 199 | 2.158 | 0.2898 | Yes |
| 6 | GRM5 | 330 | 1.836 | 0.3292 | Yes |
| 7 | HOMER2 | 371 | 1.778 | 0.3721 | Yes |
| 8 | GRIN2B | 501 | 1.621 | 0.4061 | Yes |
| 9 | GRM7 | 1018 | 1.254 | 0.4092 | Yes |
| 10 | GRIN2A | 1029 | 1.249 | 0.4403 | Yes |
| 11 | HTR3D | 1306 | 1.128 | 0.4536 | Yes |
| 12 | CBLN2 | 1319 | 1.126 | 0.4815 | Yes |
| 13 | GRIN3A | 1726 | 1.002 | 0.4843 | Yes |
| 14 | LRRTM1 | 1742 | 0.997 | 0.5088 | Yes |
| 15 | NLGN3 | 1757 | 0.994 | 0.5332 | Yes |
| 16 | DLGAP1 | 2153 | 0.892 | 0.5339 | Yes |
| 17 | HOMER3 | 2414 | 0.837 | 0.5406 | Yes |
| 18 | CACNG8 | 3242 | 0.696 | 0.5122 | No |
| 19 | SHISA9 | 3913 | 0.606 | 0.4903 | No |
| 20 | NETO2 | 4009 | 0.594 | 0.5001 | No |
| 21 | GRM1 | 4054 | 0.589 | 0.5126 | No |
| 22 | GRIA2 | 4563 | 0.537 | 0.4980 | No |
| 23 | SYNGAP1 | 4861 | 0.511 | 0.4945 | No |
| 24 | SHANK2 | 5253 | 0.475 | 0.4848 | No |
| 25 | DLG4 | 5296 | 0.472 | 0.4944 | No |
| 26 | SHANK3 | 5513 | 0.454 | 0.4939 | No |
| 27 | CACNG5 | 5826 | 0.431 | 0.4875 | No |
| 28 | GRM8 | 5877 | 0.428 | 0.4956 | No |
| 29 | GRIA1 | 6130 | 0.410 | 0.4919 | No |
| 30 | OLFM2 | 6312 | 0.399 | 0.4920 | No |
| 31 | GRIK2 | 6709 | 0.374 | 0.4794 | No |
| 32 | HTR3C | 6806 | 0.367 | 0.4834 | No |
| 33 | GRID1 | 6829 | 0.366 | 0.4915 | No |
| 34 | GRM4 | 7774 | 0.314 | 0.4469 | No |
| 35 | NLGN2 | 8355 | 0.284 | 0.4219 | No |
| 36 | HTR3E | 9021 | 0.256 | 0.3913 | No |
| 37 | DLGAP4 | 9171 | 0.250 | 0.3894 | No |
| 38 | HTR3A | 9366 | 0.241 | 0.3847 | No |
| 39 | CNIH3 | 9596 | 0.232 | 0.3779 | No |
| 40 | GRIN2C | 9894 | 0.221 | 0.3670 | No |
| 41 | NLGN1 | 10130 | 0.213 | 0.3593 | No |
| 42 | CDH9 | 10707 | 0.193 | 0.3321 | No |
| 43 | DLGAP3 | 11013 | 0.182 | 0.3198 | No |
| 44 | SHISA6 | 11953 | 0.153 | 0.2714 | No |
| 45 | GRIN1 | 11975 | 0.152 | 0.2741 | No |
| 46 | VWC2L | 12076 | 0.150 | 0.2723 | No |
| 47 | HOMER1 | 12177 | 0.147 | 0.2705 | No |
| 48 | CBLN4 | 12729 | 0.133 | 0.2432 | No |
| 49 | CNIH2 | 12879 | 0.130 | 0.2382 | No |
| 50 | DLG3 | 12968 | 0.127 | 0.2366 | No |
| 51 | GRIN2D | 13042 | 0.126 | 0.2357 | No |
| 52 | NRXN1 | 13339 | 0.118 | 0.2222 | No |
| 53 | CHRM3 | 13390 | 0.116 | 0.2224 | No |
| 54 | LHFPL4 | 13475 | 0.114 | 0.2206 | No |
| 55 | SLITRK5 | 13477 | 0.114 | 0.2234 | No |
| 56 | CHRM5 | 13765 | 0.107 | 0.2102 | No |
| 57 | CACNG2 | 14034 | 0.102 | 0.1978 | No |
| 58 | GRIN3B | 14410 | 0.093 | 0.1793 | No |
| 59 | HTR3B | 14439 | 0.093 | 0.1801 | No |
| 60 | CBLN1 | 14466 | 0.092 | 0.1810 | No |
| 61 | SHISA7 | 15072 | 0.080 | 0.1494 | No |
| 62 | DLG2 | 15075 | 0.080 | 0.1513 | No |
| 63 | GRID2 | 15087 | 0.080 | 0.1527 | No |
| 64 | CNTN4 | 15199 | 0.078 | 0.1485 | No |
| 65 | NLGN4X | 15345 | 0.075 | 0.1423 | No |
| 66 | CLSTN3 | 15630 | 0.069 | 0.1283 | No |
| 67 | TJAP1 | 15774 | 0.066 | 0.1220 | No |
| 68 | GRIA4 | 16055 | 0.059 | 0.1079 | No |
| 69 | NRXN2 | 16154 | 0.056 | 0.1039 | No |
| 70 | NETO1 | 16738 | 0.034 | 0.0723 | No |
| 71 | GRIK4 | 17392 | -0.000 | 0.0360 | No |
| 72 | GRIK3 | 17514 | -0.000 | 0.0292 | No |
| 73 | GRIK1 | 17623 | -0.000 | 0.0232 | No |